Basic Data Manipulations
Functions for Vectors and DF
| Function | Detail |
|---|---|
range() |
returns a vector containing the min and max of all the given arguments |
summary() |
produces result summaries of data stored in the vector |
unique() |
remove duplicates in the array |
table() |
build a contingency table of the counts at each combination of factor levels |
sort() |
sort a vector |
order() |
order a vector |
Transformation of V/DFs
Suppose we want to perform some operations on a dataframe. This can
be done using the function with():
d = data.frame(id = 1:6
, type = c(rep("T",3),rep("U",3)),
score = runif(6))
with( d, floor(id/score) )## [1] 1 5 3 10 6 9We can make different calculations on a dataframe in one step using
transform()
transform(d, useless = floor(id/score) , type = sample(type))## id type score useless
## 1 1 T 0.8658053 1
## 2 2 U 0.3343192 5
## 3 3 U 0.9349960 3
## 4 4 U 0.3882944 10
## 5 5 T 0.7960512 6
## 6 6 T 0.6244655 9Apply a function
The lapply() and sapply() are useful for
working with lists and dataframes. Consider
l = list( 1:5
, c("a","b")
, c(T,F,T,T) )
# apply length to the list elements
lapply(l, length)## [[1]]
## [1] 5
##
## [[2]]
## [1] 2
##
## [[3]]
## [1] 4The function length() is applied to each element of a
list and the result is also a list.
Instead, the function sapply() turn the results into a
matrix/vector if possible:
sapply(l,length)## [1] 5 2 4Functions for DF
| Function | Detail |
|---|---|
pairs() |
A quick graphical overview by the scatterplot matrix. Variables are plotted against each other |
xtabs() |
Cross-classifies variables that counts how often a combination of their levels occur |
subset() |
Return subsets of vectors, matrices or data frames which meet conditions |
pairs()
A quick graphical overview by the scatterplot matrix. Variables are plotted against each other.
The ij-th scatterplot contains x[,i] plotted
against x[,j].
d$value=with( d, floor(id/score) )
str(d)## 'data.frame': 6 obs. of 4 variables:
## $ id : int 1 2 3 4 5 6
## $ type : chr "T" "T" "T" "U" ...
## $ score: num 0.866 0.334 0.935 0.388 0.796 ...
## $ value: num 1 5 3 10 6 9## quick graphical overview by the scatterplot matrix
pairs(d[,c("id", "score","value")]
, lower.panel = panel.smooth
, upper.panel = NULL)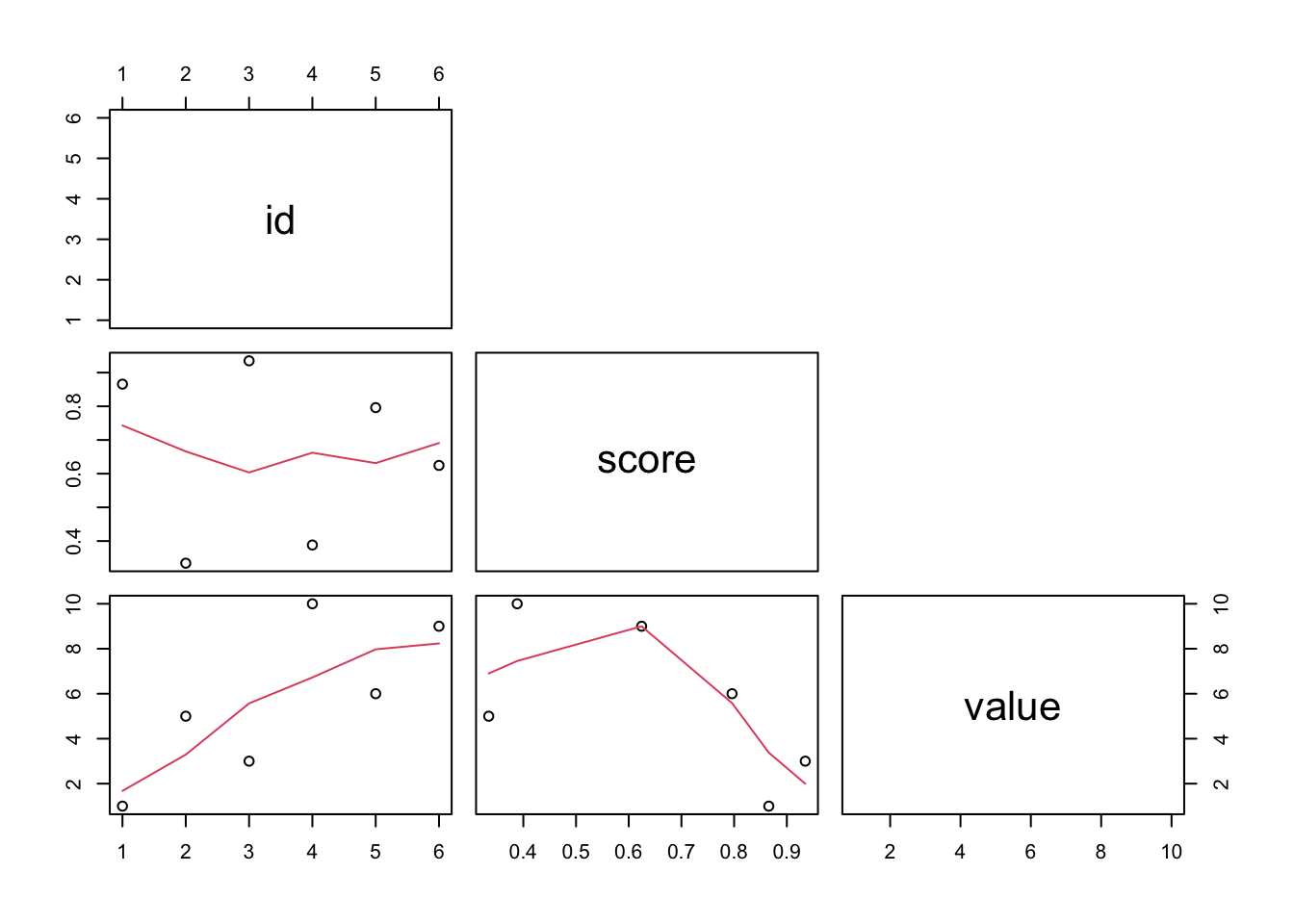
## can make my own panel to show Pearson's Correlation Coefficient (PCC)
panel.cor <- function(x, y, digits = 2, prefix = "", cex.cor, ...){
usr <- par("usr"); on.exit(par(usr))
par(usr = c(0, 1, 0, 1))
r <- abs(cor(x, y))
txt <- format(c(r, 0.123456789), digits = digits)[1]
txt <- paste0(prefix, txt)
if(missing(cex.cor)) cex.cor <- 0.8/strwidth(txt)
text(0.5, 0.5, txt, cex = cex.cor * r)
}
pairs(d[,c("id", "score","value")]
, lower.panel = panel.smooth
, upper.panel = panel.cor) # <-- here!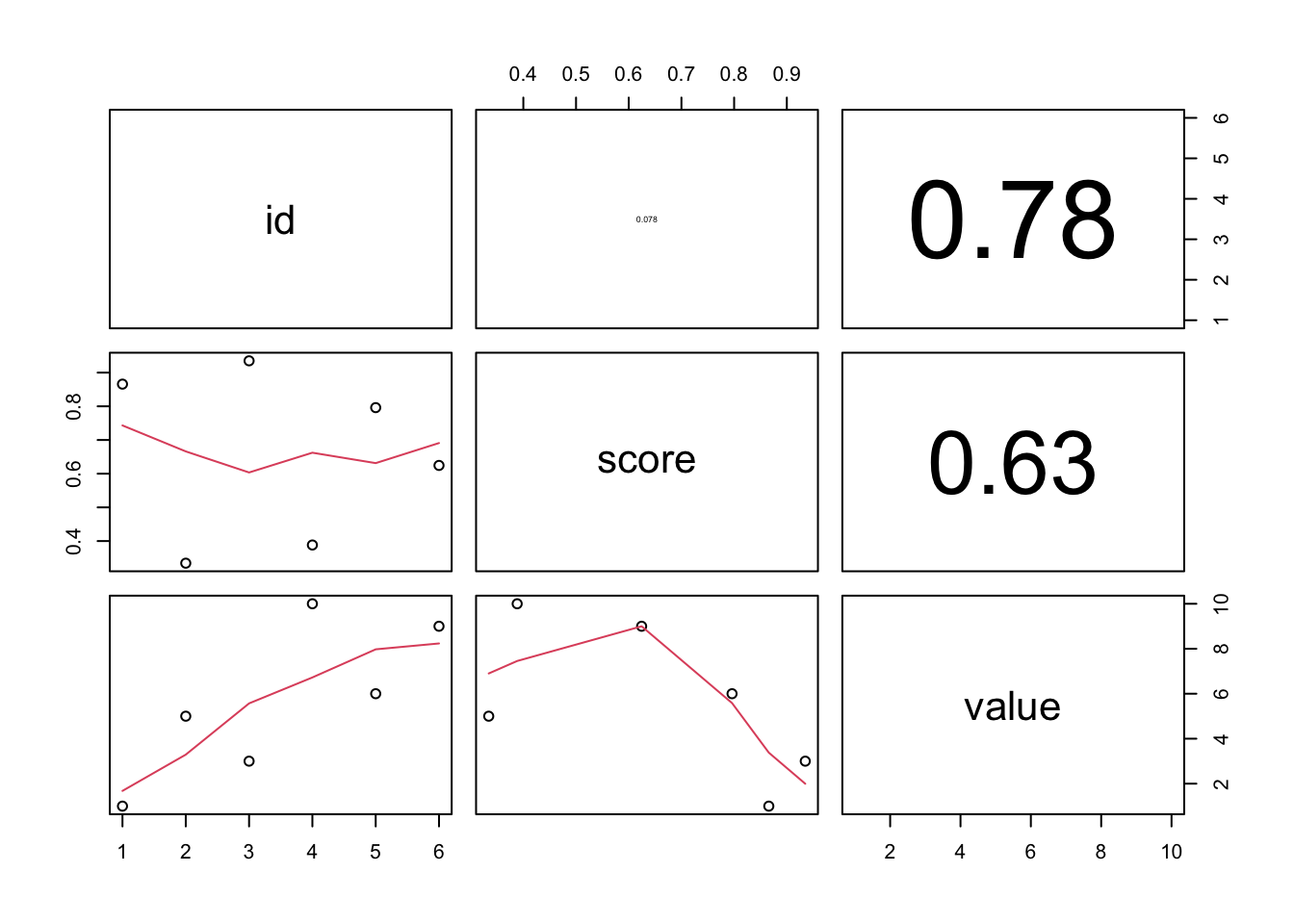
xtabs()
Cross-classifies variables counting how often a combination of their levels occur.
str(d)## 'data.frame': 6 obs. of 4 variables:
## $ id : int 1 2 3 4 5 6
## $ type : chr "T" "T" "T" "U" ...
## $ score: num 0.866 0.334 0.935 0.388 0.796 ...
## $ value: num 1 5 3 10 6 9xtabs(~value+type,d)## type
## value T U
## 1 1 0
## 3 1 0
## 5 1 0
## 6 0 1
## 9 0 1
## 10 0 1subset()
Return subsets of vectors, matrices or data frames which meet conditions
subset(d, subset = type == "T" & score > 0.5,
select = c(id, value)
)## id value
## 1 1 1
## 3 3 3Comparisons
To use filtering effectively, you have to know how to select the observations that you want using the comparison operators.
R provides the standard suite: >, >=,
<, <=, != (not equal), and
== (equal).
 Easy mistakes:
Easy mistakes:
When you’re starting out with R, the easiest mistake to make is to use
=instead of==when testing for equality.subset(d,type = "T")## id type score value ## 1 1 T 0.8658053 1 ## 2 2 T 0.3343192 5 ## 3 3 T 0.9349960 3 ## 4 4 U 0.3882944 10 ## 5 5 U 0.7960512 6 ## 6 6 U 0.6244655 9There’s another common problem you might encounter when using
==: floating point numbers. These results might surprise you!sqrt(2) ^ 2 == 2## [1] FALSE1/49 * 49 == 1
Computers use finite precision arithmetic (they obviously can’t store an infinite number of digits!) so remember that every number you see is an approximation.## [1] FALSE
Logical operators
For different types of comparisons/combinations, R provices Boolean
operators: & is “and”, | is “or”, and
! is “not”.
Figure shows the complete set of Boolean operations. x is the left-hand circle, y is the right-hand circle, and the shaded region show which parts each operator selects.
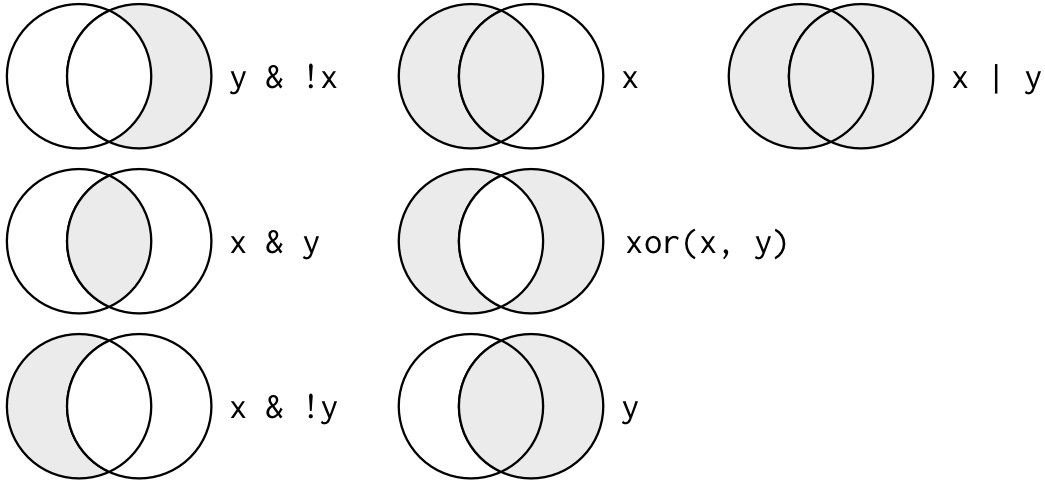
A useful short-hand for this problem is x %in% y. This
will select every row where x is one of the values in
y. We could use it to rewrite the code above:
subset(d, type %in% c("T","F"))Comparing NAs
One important feature of R that can make comparison tricky are
missing values, or NAs (“not availables”).

NArepresents an unknown value so missing values are “contagious”: Almost any operation involving an unknown value will also be unknown.
NA > 5## [1] NA10 == NA## [1] NANA + 10## [1] NANA / 2## [1] NAIf you want to determine if a value is missing, use
is.na():
is.na(c(1,NA))## [1] FALSE TRUEFunctions for Matrices
| Function | Detail |
|---|---|
scale() |
To center all columns in a matrix to have mean 0 and to rescale the columns to have variance 1 against each other |
sweep() |
Return an array obtained from an input array by sweeping out a summary statistic |
scale()
Center all columns in a matrix to have mean=0 and variance=1
If center is TRUE then centering is done by subtracting the column means (omitting NAs) of x from their corresponding columns.
m = matrix(round(runif(9),2),nr=3,nc=3)
m## [,1] [,2] [,3]
## [1,] 0.31 0.31 0.33
## [2,] 0.07 0.43 0.04
## [3,] 0.08 0.42 0.08#scaling
scale(m, center = TRUE, scale = TRUE)## [,1] [,2] [,3]
## [1,] 1.1539172 -1.1514402 1.1453126
## [2,] -0.6137858 0.6508140 -0.6999132
## [3,] -0.5401315 0.5006262 -0.4453993
## attr(,"scaled:center")
## [1] 0.1533333 0.3866667 0.1500000
## attr(,"scaled:scale")
## [1] 0.13576941 0.06658328 0.15716234sweep()
Return an array obtained from an input array by sweeping out a summary statistic.
“clean an area thoroughly by brushing away all dirt or litter”
str(m)## num [1:3, 1:3] 0.31 0.07 0.08 0.31 0.43 0.42 0.33 0.04 0.08# median value of each row of the matix
row.med <- apply(m, MARGIN = 1, FUN = median)
row.med## [1] 0.31 0.07 0.08# subtracting the median value of each row
sweep(m, MARGIN = 1, STATS = row.med, FUN = "-")## [,1] [,2] [,3]
## [1,] 0 0.00 0.02
## [2,] 0 0.36 -0.03
## [3,] 0 0.34 0.00A work by Matteo Cereda and Fabio Iannelli