Regular Expressions
A regular expression (regex) is a sequence of characters that define a search pattern.
Regex Syntax
To read more about the specifications and technicalities of regex in
R you can find help at help(regex) or
help(regexp)
Metacharacters
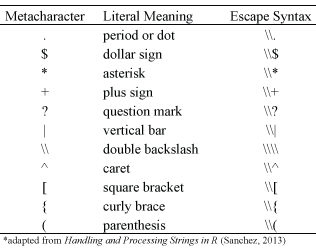
Metacharacters consist of NON-alphanumeric symbols, as:
. \\\ | ( ) [ { $ * + ?To match metacharacters in R you need to escape them with a double backslash
\\.
To find and replace metacharacters we can use the function
sub() and gsub()
# substitute $ with !
sub(pattern = "\\$", "\\!", "I love R$")## [1] "I love R!"# substitute ^ with carrot
sub(pattern = "\\^", "carrot", "My daughter has a ^ with almost every meal!")## [1] "My daughter has a carrot with almost every meal!"# substitute \\ with whitespace
gsub(pattern = "\\\\", " ", "I\\need\\space")## [1] "I need space"Sequences
To match a sequence of characters we can apply short-hand notation which captures the fundamental types of sequences.
The following displays the general syntax for these common sequences:

# substitute any digit with an underscore
gsub(pattern = "\\d", "_", "I'm working in RStudio v.0.99.484")## [1] "I'm working in RStudio v._.__.___"# substitute any non-digit with an underscore
gsub(pattern = "\\D", "_", "I'm working in RStudio v.0.99.484")## [1] "_________________________0_99_484"# substitute any whitespace with underscore
gsub(pattern = "\\s", "_", "I'm working in RStudio v.0.99.484")## [1] "I'm_working_in_RStudio_v.0.99.484"# substitute any wording with underscore
gsub(pattern = "\\w", "_", "I'm working in RStudio v.0.99.484")## [1] "_'_ _______ __ _______ _._.__.___"Character classes
To match one of several characters in a specified set we can enclose the characters of concern with square brackets
[ ]
To match any characters not in a specified character set we can
include the caret ^at the beginning of the set within the
brackets.

To find character classes we can use the function
grep()
x <- c("RStudio", "v.0.99.484", "2015", "09-22-2015", "grep vs. grepl")
# find any strings with numeric values between 0-9
grep(pattern = "[0-9]", x, value = TRUE)## [1] "v.0.99.484" "2015" "09-22-2015"# find any strings with numeric values between 6-9
grep(pattern = "[6-9]", x, value = TRUE)## [1] "v.0.99.484" "09-22-2015"# find any strings with the character R or r
grep(pattern = "[Rr]", x, value = TRUE)## [1] "RStudio" "grep vs. grepl"# find any strings that have non-alphanumeric characters
grep(pattern = "[^0-9a-zA-Z]", x, value = TRUE)## [1] "v.0.99.484" "09-22-2015" "grep vs. grepl"POSIX character classes
Closely related to regexp character classes are POSIX (Portable
Operating System Interface) character classes which are expressed
in double brackets [[ ]].
POSIX is an IEEE standard designed to facilitate application portability.

x <- "I like beer! #beer, @wheres_my_beer, I like R (v3.2.2) #rrrrrrr2015"
# remove space or tabs
gsub(pattern = "[[:blank:]]", replacement = "", x)## [1] "Ilikebeer!#beer,@wheres_my_beer,IlikeR(v3.2.2)#rrrrrrr2015"# replace punctuation with whitespace
gsub(pattern = "[[:punct:]]", replacement = " ", x)## [1] "I like beer beer wheres my beer I like R v3 2 2 rrrrrrr2015"# remove alphanumeric characters
gsub(pattern = "[[:alnum:]]", replacement = "", x)## [1] " ! #, @__, (..) #"Quantifiers
When we want to match a certain number of characters that meet a certain criteria we can apply quantifiers to our pattern searches.
The quantifiers we can use are:

# match states that contain z
grep(pattern = "z+", state.name, value = TRUE)## [1] "Arizona"# match states with two s
grep(pattern = "s{2}", state.name, value = TRUE)## [1] "Massachusetts" "Mississippi" "Missouri" "Tennessee"# match states with one or two s
grep(pattern = "s{1,2}", state.name, value = TRUE)## [1] "Alaska" "Arkansas" "Illinois" "Kansas" "Louisiana"
## [6] "Massachusetts" "Minnesota" "Mississippi" "Missouri" "Nebraska"
## [11] "New Hampshire" "New Jersey" "Pennsylvania" "Rhode Island" "Tennessee"
## [16] "Texas" "Washington" "West Virginia" "Wisconsin"Pattern Finding Functions
| Function | Task |
|---|---|
grep() |
find a pattern in a character vector |
grepl() |
find position in a character vector matching a pattern |
regexpr() |
find exactly where the pattern exists |
grep()
To find a pattern in a character vector and to have the element
values or indices as the output use grep():
# use the built in data set `state.division`
head(as.character(state.division))## [1] "East South Central" "Pacific" "Mountain" "West South Central"
## [5] "Pacific" "Mountain"# find the elements which match the patter
grep("North", state.division)## [1] 13 14 15 16 22 23 25 27 34 35 41 49# use 'value = TRUE' to show the element value
grep("North", state.division, value = TRUE)## [1] "East North Central" "East North Central" "West North Central" "West North Central"
## [5] "East North Central" "West North Central" "West North Central" "West North Central"
## [9] "West North Central" "East North Central" "West North Central" "East North Central"# can use the 'invert' argument to show the non-matching elements
grep("North | South", state.division, invert = TRUE)## [1] 2 3 5 6 7 8 9 10 11 12 19 20 21 26 28 29 30 31 32 33 37 38 39 40 44 45 46 47 48
## [30] 50grepl()
To find a pattern in a character vector and to have logical
(TRUE/FALSE) outputs use grepl():
grepl("North | South", state.division)## [1] TRUE FALSE FALSE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE TRUE
## [15] TRUE TRUE TRUE TRUE FALSE FALSE FALSE TRUE TRUE TRUE TRUE FALSE TRUE FALSE
## [29] FALSE FALSE FALSE FALSE FALSE TRUE TRUE TRUE FALSE FALSE FALSE FALSE TRUE TRUE
## [43] TRUE FALSE FALSE FALSE FALSE FALSE TRUE FALSEregexpr()
To find exactly where the pattern exists in a string use
regexpr():
x <- c("v.111", "0v.11", "00v.1", "000v.", "00000")
regexpr("v.", x)## [1] 1 2 3 4 -1
## attr(,"match.length")
## [1] 2 2 2 2 -1
## attr(,"index.type")
## [1] "chars"
## attr(,"useBytes")
## [1] TRUEThe first element provides the starting position of the match in each element.
The value -1 means there is no match.
The second element (attribute “match length”) provides the length of the match.
The third element (attribute “useBytes”) has a value TRUE meaning matching was done byte-by-byte rather than character-by-character.
Pattern Replacement Functions
| Function | Task |
|---|---|
sub() |
replace the first matching occurrence of a patter |
gsub() |
replace all matching occurrences of a pattern |
sub()
To replace the first matching occurrence of a pattern use ‘sub()’:
new <- c("New York", "new new York", "New New New York")
new## [1] "New York" "new new York" "New New New York"# Default is case sensitive
sub("New", replacement = "Old", new)## [1] "Old York" "new new York" "Old New New York"# use 'ignore.case = TRUE' to perform the obvious
sub("New", replacement = "Old", new, ignore.case = TRUE)## [1] "Old York" "Old new York" "Old New New York"gsub()
To replace all matching occurrences of a pattern use
gsub():
gsub("New", replacement = "Old", new)## [1] "Old York" "new new York" "Old Old Old York"gsub("New", replacement = "Old", new, ignore.case = TRUE)## [1] "Old York" "Old Old York" "Old Old Old York"Splitting Character Vectors
To split the elements of a character string use
strsplit():
x <- paste(state.name[1:10], collapse = " ")
# output will be a list
strsplit(x, " ")## [[1]]
## [1] "Alabama" "Alaska" "Arizona" "Arkansas" "California" "Colorado"
## [7] "Connecticut" "Delaware" "Florida" "Georgia"# output as a vector rather than a list
unlist(strsplit(x, " "))## [1] "Alabama" "Alaska" "Arizona" "Arkansas" "California" "Colorado"
## [7] "Connecticut" "Delaware" "Florida" "Georgia"Usefull links
The RegExr web tool https://regexr.com/
A RegExp cheatsheet https://www.rstudio.com/wp-content/uploads/2016/09/RegExCheatsheet.pdf
A work by Matteo Cereda and Fabio Iannelli